Bioinformatics as applied to the study of genome structure and function
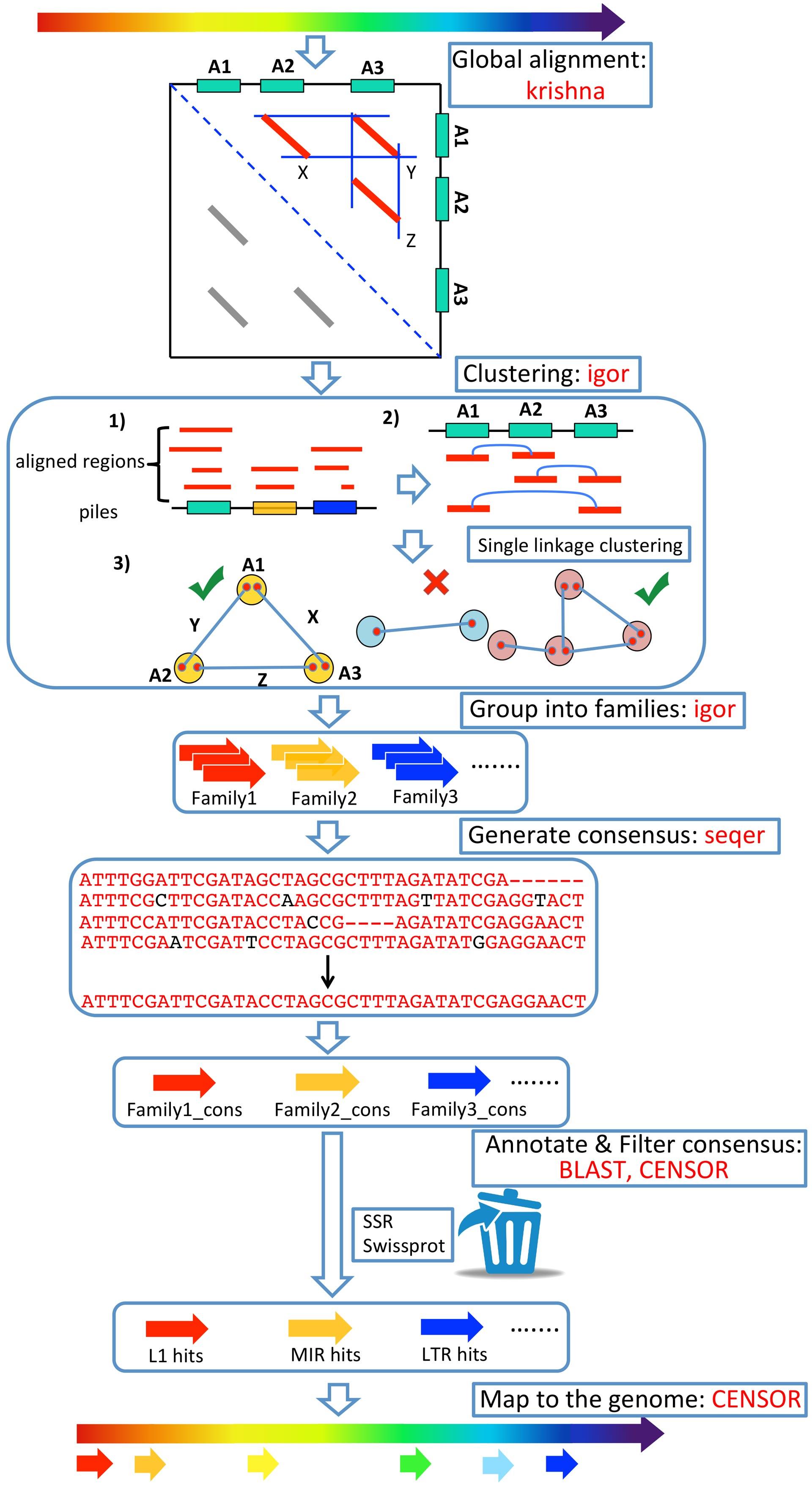
Fig 1. Comprehensive ab initio Repeat Pipeline (CARP). Refer to paper for more information.
Larger image
Expand your knowledge in bioinformatics and computational genetics. Study an honours research project in molecular and biomedical science.
Our research has applications in biomedical science (cancer, aging), agriculture (plants and animals) and is driven by understanding fundamental aspects of how genomes evolve through the amplification and horizontal transfer of transposable elements. My lab has pioneered the identification of horizontal transfer of retrotransposons.
Prerequisites:
- It would be helpful (but not required) if you already had some familiarity with either Unix/Linux command line or computer programming and a reasonable grasp of statistics.
- All that is really required is curiosity and an interest in bioinformatics.
For specific projects, I prefer to identify students' interests through a brief discussion with them and then suggest a project that incorporates their interests with an area of our research.
Bioinformatics projects
Honours projects in my group are available in the following areas:
| Creating new software tools for biologists to analyse genomic data | The first involves creating new methods and implementing them as software to allow discovery and annotation of transposable elements in newly sequenced genomes, and development of methods to identify somatic mutations caused by transposons. |
| Analysing genome data using computational methods | The second involves applying our software tools using all available public genome sequence assemblies (currently almost 1000 available for metazoans) for evidence of novel transposons, transposon horizontal transfer and how transposons contribute to somatic genome structural variation. |
| Detection and annotation of transposons in genome sequences/assemblies | The third honours project has a goal to improve the current software/workflow pipeline for the detection and annotation of novel transposable elements in genome sequences. The current pipeline has limitations for the automated characterisation of certain classes of transposable elements that need to be addressed. This paper provides a description of the existing pipeline. |

Researchers of the Centre for Bioinformatics and Computational Genetics
Supervisors
Co-supervisors: Dr Stephen Pederson | Dr Dan Kortschak | Dr Nathan Watson-Haigh | Dr Jimmy Breen | Associate Professor Gary Glonek
Research area: Centre for Bioinformatics and Computational Genetics
Recommended honours enrolment: Honours in Molecular and Biomedical Science
