Cellular-stress induced activation of non-TFEB regulated lysosomal genes
This molecular and biomedical science project will investigate the role of the eIF2α/ATF4 pathway in stress-induced transcription of non-TFEB genes.
The ability to catabolise and recycle molecules that are detrimental to its function is crucial for cell survival [1]. Lysosome biogenesis (formation of new lysosomes, the organelle primarily responsible for the degradation of biomolecules) ensures that this occurs.
Lysosomal dysfunction is linked to a range of diseases, including neurodegenerative disorders such as Alzheimer’s Disease and is also important in cancer. Thus, a full understanding of the control of lysosomal gene expression is essential.
Lysosomes are generated at a steady rate in normal conditions. However, during cellular insults (such as build-up of toxic products, or nutrient depletion) the expression of lysosomal genes increases, as do the number and size of lysosomes. Cellular stress-induced transactivation of most lysosomal genes is mediated by the transcription factor EB (TFEB) [2]. Normally, TFEB resides in the cytoplasm in an inactive form; however, during cellular stress, TFEB translocates to the nucleus, allowing it to drive the expression of its target genes. The DNA sequence to which TFEB binds is known as the Coordinated Lysosomal Expression and Regulation (CLEAR) element [2; 3].
Autophagy (the regulated destruction of cellular molecules and structures) is a process that is intricately linked with lysosomal biogenesis and function, and a cohort of autophagy related genes is also transactivated by TFEB during cellular stress [3].
The cellular insults that activate TFEB also increase the expression of lysosomal genes that are not regulated by TFEB, although the means by which this occurs remain elusive. One mechanism which could be involved is the eukaryotic initiation factor (eIF) 2α/activating transcription factor (ATF) 4 pathway.
Here, in cases of amino acid deprivation or endoplasmic reticulum stress, eIF2α is phosphorylated by a specific kinase (GCN2 or PERK), which in turn increases levels of ATF4, a transcription factor that, directly or indirectly, increases the transcription of a subset of genes implicated in autophagy [4;5].
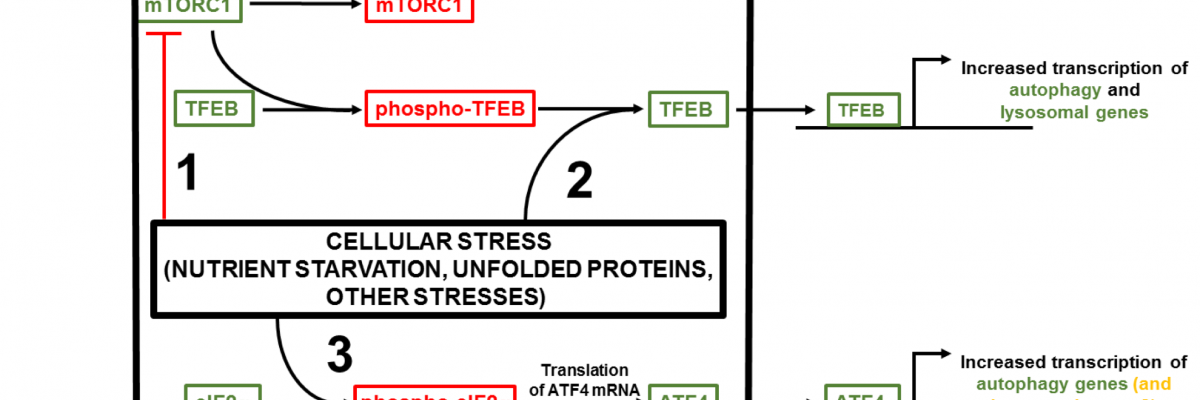
Cellular stress-induced transactivation of autophagy and lysosomal genes. In normal conditions, transcription factor EB (TFEB) is kept in the cytoplasm in an inactive form.
Hypothesis
The eIF2α/ATF4 pathway contributes to controlling lysosomal gene expression.
Aim
To investigate the role of the eIF2α/ATF4 pathway in stress-induced transcription of non-TFEB genes.
Cellular stress-induced transactivation of autophagy and lysosomal genes. In normal conditions, transcription factor EB (TFEB) is kept in the cytoplasm in an inactive form.
This is elicited by the kinase activity of mechanistic target of rapamycin 1 (mTORC1), which is tethered to the lysosomal membrane on the cytoplasmic side. In conditions of cellular stress, such as nutrient starvation, mTORC1 is released from the lysosomal membrane and cannot phosphorylate TFEB (1). Furthermore, phosphatase activity toward any already phosphorylated TFEB is enhanced (2). These processes ensure efficient translocation of TFEB to the nucleus, thus increasing expression of target autophagy and lysosomal genes. A cohort of autophagy genes is regulated by the eukaryotic initiation factor (eIF) 2α/activating transcription factor (ATF) 4 pathway.
During cellular stress, eIF2α is phosphorylated (3), which, while decreasing the translation of most mRNAs, increases the translation of a select few, including ATF4, a transcription factor that increases transcription of a distinct subset of genes implicated in autophagy. This project will address the question ‘could this pathway also be involved in the transcription of lysosomal genes that are not regulated by TFEB?’ Red/green refers to inactive/active components.
Project outline
Initially, human cell lines (including 293T and A549) and mouse embryonic fibroblasts (MEFs) will be treated with pharmacological activators of the eIF2α/ATF4 pathway. These include brefeldin A (which promotes endoplasmic reticulum stress) and histidinol (which mimics amino acid starvation). Both can induce expression of ATF4. Quantitative real-time PCR (qRT-PCR) and/or immunoblotting will be then be used to determine the levels of candidate lysosomal mRNA and proteins, respectively. As controls, levels of ATF4 and C/EBP homologous transcription factor (CHOP, a direct target of ATF4) will concurrently be determined, as these are expected to increase in these circumstances.
To further confirm the responses to the chemical treatments, the previous experiments will be repeated using MEFs where stress-inducible eIF2α phosphorylation has been genetically inactivated by knocking out GCN2 or PERK. If levels of a candidate ATF4-regulated lysosomal gene are increased by brefeldin A or histidinol in normal cells, but not in the genetically modified lines, this will further implicate the eIF2α/ATF4 pathway in non-TFEB driven lysosomal gene expression.
Luciferase reporter assays will then be used to delineate the specific DNA sequence (known as the response element) to which the trans-activating protein associates. Finally, to identify which candidate transcription factors (such as ATF4 or CHOP) bind to the response elements of the genes of interest, chromatin immunoprecipitation (ChIP) assays will be employed.
In this project, you will learn a range of up-to-date molecular biology techniques and the data will contribute to understanding the control of autophagy in relation to human disease.
Key references
- Ballabio A. 2016. EMBO Mol Med 8: 73-76.
- Sardiello M, et al. 2009. Science 325: 473-477.
- Palmieri M, et al. 2011. Hum Mol Genet 20: 3852-3866.
- B’chir W, et al. 2013. Nucl Acids Res 41: 7683-7699.
- Han J, et al. 2013. Nat Cell Biol 15: 481-490.

Supervisor
Research area: Cell signalling and gene regulation; Nutrition and metabolism, South Australian Health and Medical Research Institute
Recommended honours enrolment: Honours in Molecular and Biomedical Science
