Role of eEF2K & cancer cell migration & invasion
Study cell signalling and gene regulation through the exploration of the role of eukaryotic elongation factor 2 kinase (eEF2K) and cancer cell migration and invasion.
Tumour metastasis is a hallmark of malignant carcinomas and is associated with poor prognosis and survival rates among high-risk cancer patients. Most cancer patients die of the resulting secondary tumours rather than the primary one.
According to the US National Cancer Institute Surveillance, Epidemiology and End Results (SEER) registries, in 2016, the average survival rates from diagnosed metastatic cancer patients are only 25% or less compared to patients suffering localised (organ-defined) or regional (lymph node invasion) cancers. This has not improved much during the last decade.
In recent years, regulatory mechanisms driven by diverse molecular signalling pathways controlling protein synthesis have been revealed to play central roles in tumour metastasis and progression. Thus, the capacity of the translation machinery of cancer cells to “fine-tune” gene expression has attracted increasing attention in both basic and translational cancer research fields.
Global protein synthesis is generally increased in cancer cells, and accumulating data indicate that the translational regulatory networks somehow steer events to adapt to microenvironmental stress conditions and satisfy the survival and spreading requirements of such cells.
Regulation of mRNA translation is essential for reshaping the cellular proteome to initiate tumorigenesis and metastasis. However, the underlying mechanisms are incompletely understood. Our recent data show that control of translation elongation plays roles in tumour cell migration and invasion by regulating the proteome of cancer cells.
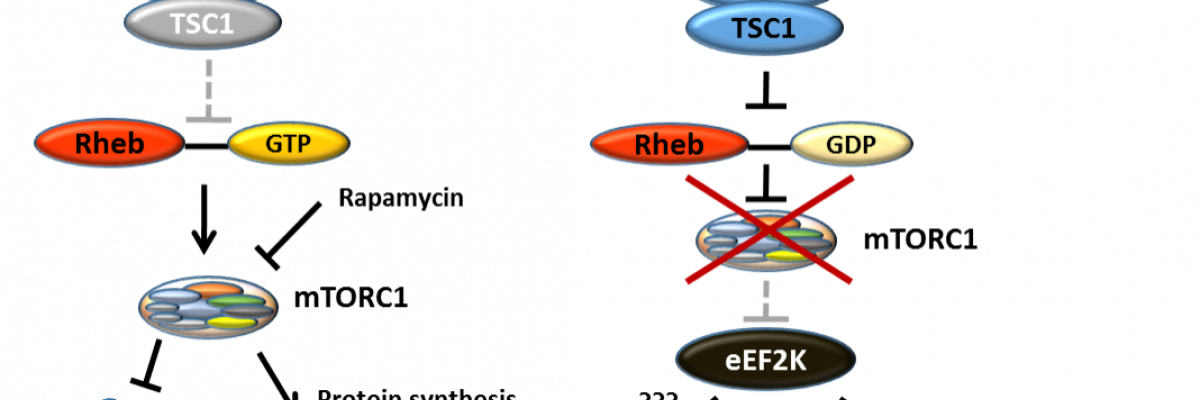
Figure 1. Cancer cells survive and gain the ability to spread under microenvironmental stress conditions through the inhibition of mTORC1 and subsequent activation of eEF2K.
Eukaryotic elongation factor 2 (eEF2) kinase (eEF2K) is a very specific, atypical protein kinase whose only known and validated substrate is eEF2. eEF2 mediates the translocation of ribosomes along mRNAs and the concomitant movement of aminoacyl-tRNA from A into P site.
Phosphorylation of eEF2 on Thr56 by eEF2K impedes the binding of eEF2 to ribosomes and hence impairs translation elongation. Interestingly, eEF2K is highly expressed in some cancers such as medulloblastoma, glioblastoma, breast cancer and pancreatic tumours. It is activated under several stress conditions typically present in the tumour microenvironment, e.g. lack of nutrients, hypoxia, acidosis and/or energy deprivation.
We have shown that eEF2K activity can regulate the expression of proteins that important in cell migration and thus tumour metastasis. eEF2K also helps cancer cells survive stress conditions such as nutrient starvation or hypoxia, perhaps by regulating cell metabolism. As it is not essential, on-target side-effects are not expected, so eEF2K may be a valuable new target for preventing cancer metastasis. We are working with drug discovery companies to study this.
Recently we have evidence that pharmacological or genetic blockage of eEF2K prevents cell migration and invasion in a variety of cancer cells including lung, breast and prostate cancer cells.
In this project, we will explore the molecular mechanisms by which eEF2K activation encourage cancer cell spreading. We will use the state-of-the-art CRISPR/Cas9 (Clustered regularly interspaced short palindromic repeats) gene editing technology to delete eEF2K in different cancer cell lines as models to explore the role of eEF2K in cancer cell biology including their migration and invasion. The migratory potential of these cells will be tested by two-dimensional wound-healing and three-dimensional transwell invasion assays.
We will study the effects of genetic deletion of eEF2K on cell growth, proliferation and cell cycle, migration, invasion, metabolism and on protein synthesis in these cancer cells. We will also compare the spectrum of proteins being synthesised in wild-type (WT) and eEF2K-/- cells, by using the BONCAT (bioorthogonal non-canonical amino acid tagging)-pSILAC (pulsed stable isotope-labelling with amino acids in cell culture) technique. These methods are already fully established in the host lab.
The data will provide new insights into the potential of eEF2K as a target in anti-cancer therapy.
Key reference: Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153: 1064-79.

Supervisor
Research area: Cell signalling and gene regulation; Nutrition and metabolism, South Australian Health and Medical Research Institute
Recommended honours enrolment: Honours in Molecular and Biomedical Science
