Synthetic phage therapy
Study honours in synthetic phage therapy and investigate the structural biology of bacteriophage proteins.
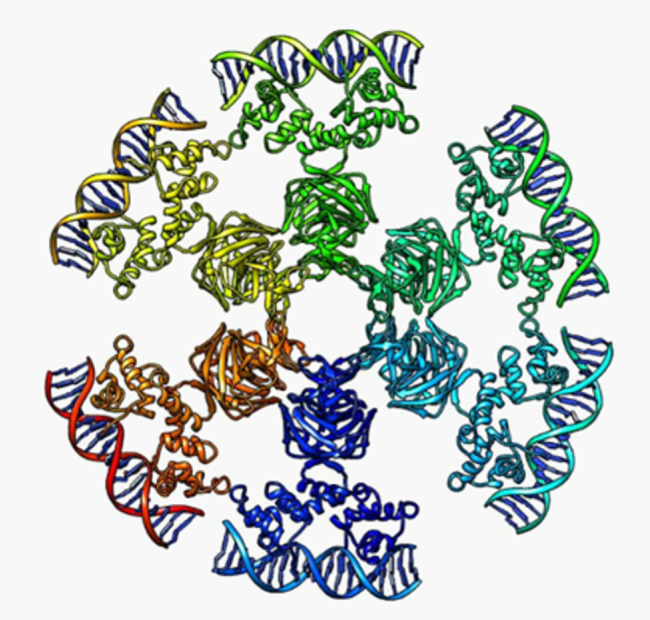
Knowing the three dimensional structure of a protein allows us to have a detailed, molecular level understanding of the function of that protein.
For bacteriophage 186, we have crystal structures of the CI repressor, which forms a wheel-like complex of 12 subunits, and of CII, which is a transcriptional activator protein able to interact with RNA polymerase.
In addition to understanding the molecular basis of phage decision making, we are also interested in developing ‘synthetic’ phage with enhanced bacterial killing properties for phage therapy.
One approach is to develop phage able to display various enzymatic activities on their surface. For example, a phage which displays an enzyme capable of breaking down biofilms might be more effective in reaching its bacterial target.
To evaluate which phage structural proteins would provide the best platform for surface display, we would like to survey a number of proteins from bacteriophage 186, and from other phage, to assess their suitability for structure determination by either X-ray crystallography or cryo-EM.
This project would involve cloning, expression and purification of some candidate proteins, followed by structure determination of the promising candidates.
Study synthetic biology, biochemistry, genetics and mathematical modelling
Our primary experimental systems are two E. coli bacteriophages, lambda and 186. These temperate phages can replicate their genomes using alternative developmental pathways, lysis and lysogeny, and are some of the simplest organisms to make developmental decisions.
Despite their relative simplicity, the phage systems combine a wide range of gene control mechanisms in complex ways and have many lessons to teach us.
Bacteriophage lambda continues to be a key model system for many molecular biological processes; phage 186 is less well characterised but provides a powerful comparison with lambda, as it achieves similar outcomes using different regulatory circuits.

Supervisors
Associate Professor Keith Shearwin
Co-supervisor: Dr Nan Hao | Dr Ian Dodd
Research area: Synthetic biology, biochemistry, genetics and mathematical modelling
Recommended honours enrolment: Honours in Molecular and Biomedical Science
