Systematic analysis of the eIF4E interactome
Conduct a systematic analysis of the eIF4E interactome to understand the distinct functional roles of the three mammalian eIF4E proteins in translation initiation.
One of the most intriguing findings to arise from the recent availability of genome-wide datasets measuring the rates of synthesis and turnover of both mRNA and protein is that cellular protein abundance is determined mainly by controls at the level of translation.
Indeed, the majority of variance (~55%) in protein levels is determined by the rates of translation of their mRNAs.
Translational control of protein amount has several advantages: for example, one, production of a given protein can be quickly activated at the ribosome, without the need to first transcribe, process and export the template mRNA, and two, a protein’s synthesis can be restricted to specific subcellular locales, which is essential in polarised cells such as neurons or (cancer) cells undergoing directed migration.
While we now understand a great deal about the general mechanisms of translation, our insights into the broader picture of how both cellular environment and sequence-specific translational regulatory mechanisms cooperate to control the translational efficiency (TE) of a given mRNA under specific states are poor.
However, it is clear that translation initiation, i.e., the step where ribosomes are recruited onto the mRNA and locate the start codon, is a key site of regulation. Initiation involves the association of the mRNA with a set of proteins termed eukaryotic initiation factors, eIFs.
There are 3 core eIFs (which together form ‘the eIF4F complex’): the mRNA 5′ cap-binding protein eIF4E (which occurs as three different forms encoded by different genes); the RNA helicase eIF4A; and the scaffold eIF4G. The eIF4F: mRNA assembly – the classic 'closed-loop' structure – is the platform onto which the 40S small ribosomal subunit, already bound to other translation factors and termed the 43S pre-initiation complex) docks, starting the process of translation.
Interestingly, while the vast majority of translation initiation research has studied the eIF4E-type protein eIF4E1, there are two other mammalian eIF4E isoforms (arising from the other two genes); we henceforth refer to them as eIF4E1, eIF4E2 and eIF4E3 (the eIF4Es). We recently made the significant discovery that eIF4E2 may act as the key mediator of a stress response that achieves selective translational inhibition of mRNAs involved in cellular growth.
As the field lacks even basic experimental data on eIF4E2 and eIF4E3, understanding our important and novel findings requires a research strategy that yields comprehensive data on the mRNA and protein cohorts of each eIF4E, and uses the data to build and test models of eIF4E function.
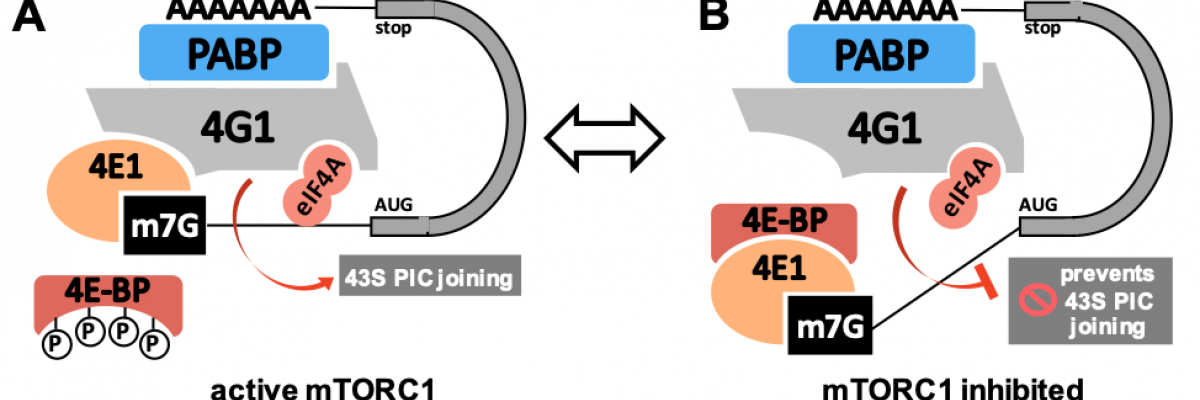
Figure 1. (A) Active mTORC1 phosphorylates 4E-BP, preventing 4E1 binding. (B) mTORC1 inhibition leads to 4E-BP hypophosphorylation; 4E-BP now binds 4E1 and blocks eIF4F.
In this project, we will probe how changes in the cellular environment affect the individual functions and macromolecular interactions of eIF4E1, eIF4E2 and eIF4E3.
In brief, we plan to investigate, using human cell lines grown in culture, how alterations to the cellular environment that have been designed to alter mTORC1 (mammalian target of rapamycin complex 1) signalling affect both the mRNA and protein partners of the three eIF4Es. As mTORC1 activity has been shown to play a major role in the regulation of translation initiation (partly by mechanisms which are well understood and partly through unknown means) we will identify the individual mRNAs and protein partners of each eIF4E under different cellular growth conditions.
Next, to understand how these cellular environments alter translation, we will use polysome profiling to determine the translational efficiency (TE) of individual mRNAs, combined with pulsed, isotopic labelling (BONCAT, see above) of cellular proteins to allow a direct readout of the synthesis rates of individual types of proteins.
Finally, by integrating the information we will obtain about the translational activity of individual mRNAs with our knowledge of each message’s eIF4E isoform association, across five different cell contexts, we will gain a number of novel and comprehensive insights into the functional roles of each eIF4E in translation initiation which can then be tested by experiment.
This project involves a range of state-of-the-art techniques.
Key references
- Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., Chen, W., and Selbach, M. (2011). Global quantification of mammalian gene expression control. Nature 473, 337-342.
- Huo, Y., Iadevaia, V., and Proud, C. G. (2011). Differing effects of rapamycin and mTOR kinase inhibitors on protein synthesis. Biochem Soc Trans 39, 446-450.

Supervisor
Research area: Cell signalling and gene regulation; Nutrition and metabolism, South Australian Health and Medical Research Institute
Recommended honours enrolment: Honours in Molecular and Biomedical Science
