Gene scans provide deep insights into plant evolution
To say that plants are important is a truism beyond doubt. They fill almost every ecosystem and niche on earth and support almost every other life form on the planet.
But, with close to half a million species out there, have you ever wondered where they come from and how such diversity evolved? Well you’ll be pleased to hear that it’s a complex and sordid tale of clonality and sex between species.
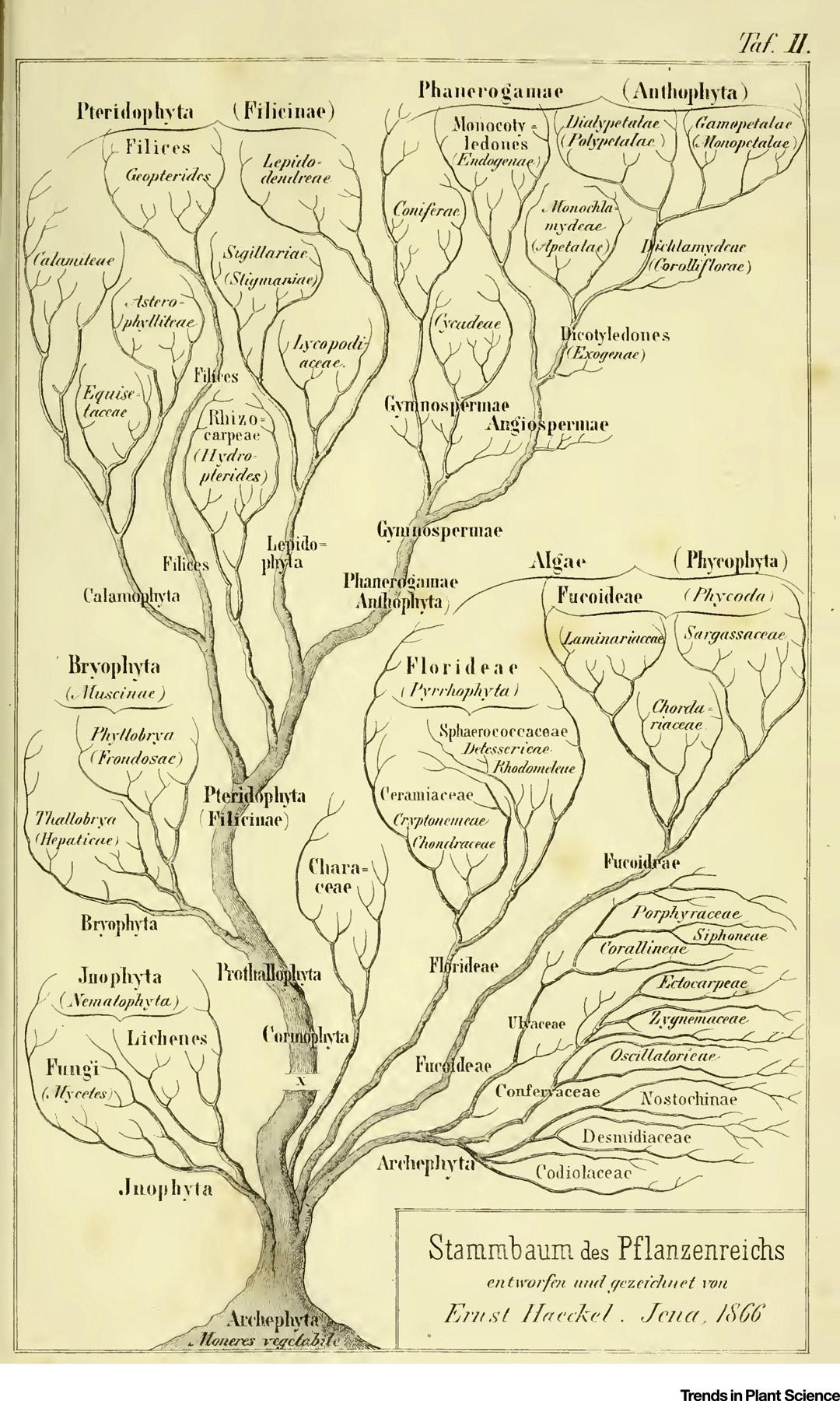
The first evolutionary tree of plants was published by the German zoologist Ernst Haeckel in his 1866 book General Morphology of Organisms.
View larger image
There have been many attempts over the year to piece together the evolutionary history of plants.
But studies that have used the structural features of plants (mostly their flowers, number of petals etc) run into problems as some structures look similar but rather than being inherited, they have evolved multiple times in different groups because the feature is useful.
More recently DNA analysis has helped shed light on the relationship between plants, but the problem has been which genome to analyse. Plants have three genomes, one in their nucleus, one in their mitochondria (the battery-like power centres of the cell) and one in their chloroplasts (the solar cell like structures that convert light into energy).
For various reasons the chloroplast genome has been the focus of most evolutionary work, but it doesn’t tell the whole evolutionary story, and particularly when hybridisation occurs between species the evolutionary picture can be difficult to piece together.
But with the cost of genomics plummeting, a group of scores of scientists from around the world (including myself - Andrew Lowe - and Ed Biffin from the State Herbarium of South Australia) participated in the One Thousand Plant Transcriptome Initiative.
Scientists sequenced over 400 groups of genes (from the nucleus, chloroplast and mitochondria) from over 1000 plant species that span the diversity of life on earth – from algae, through mosses and ferns, conifers, orchids and grasses, through tropical trees and onto roses and daisies (the most recently evolved groups).

The latest picture of plant evolution (Nature)

Gnetum gnemon (Nature)
Having sequence data from so many genes, allows the true path of evolution of a species to be traced, rather than just a single genome. Analysis of large groups of genes also allows incidents of genome duplication (also known as polyploidy) or gene family expansion to be easily identified.
So, what did we find?
Across the plant evolutionary tree of life genome duplication, periods of rapid speciation, massive expansion in the number and complexity of genes and periods of extinction have all played important roles.
In particular, a massive expansion in the number and complexity of genes predates the origin of all plants, but also land plants and vascular plants (those with stems). Other striking findings are that whole-genome duplications have occurred repeatedly throughout the evolution of flowering plants and ferns.
The availability of high-quality plant genome sequences and advances in functional genomics is revolutionising our ability to understand evolution across the tree of life, and in particularly has given us an insight into the evolutionary history of plants never before realised.
About this article
- Written by Professor Andy Lowe, this article was republished from Biodiversity Revolution: Thoughts from the vanguard of biodiversity research. View the original article.
Further reading
- One thousand plant transcriptomes and the phylogenomics of green plants, Nature volume 574.
