CRISPR genome wide screens & viral replication
Use CRISPR/Cas9 genome wide screens to identify viral host dependency factors and novel innate immune genes that control viral replication.
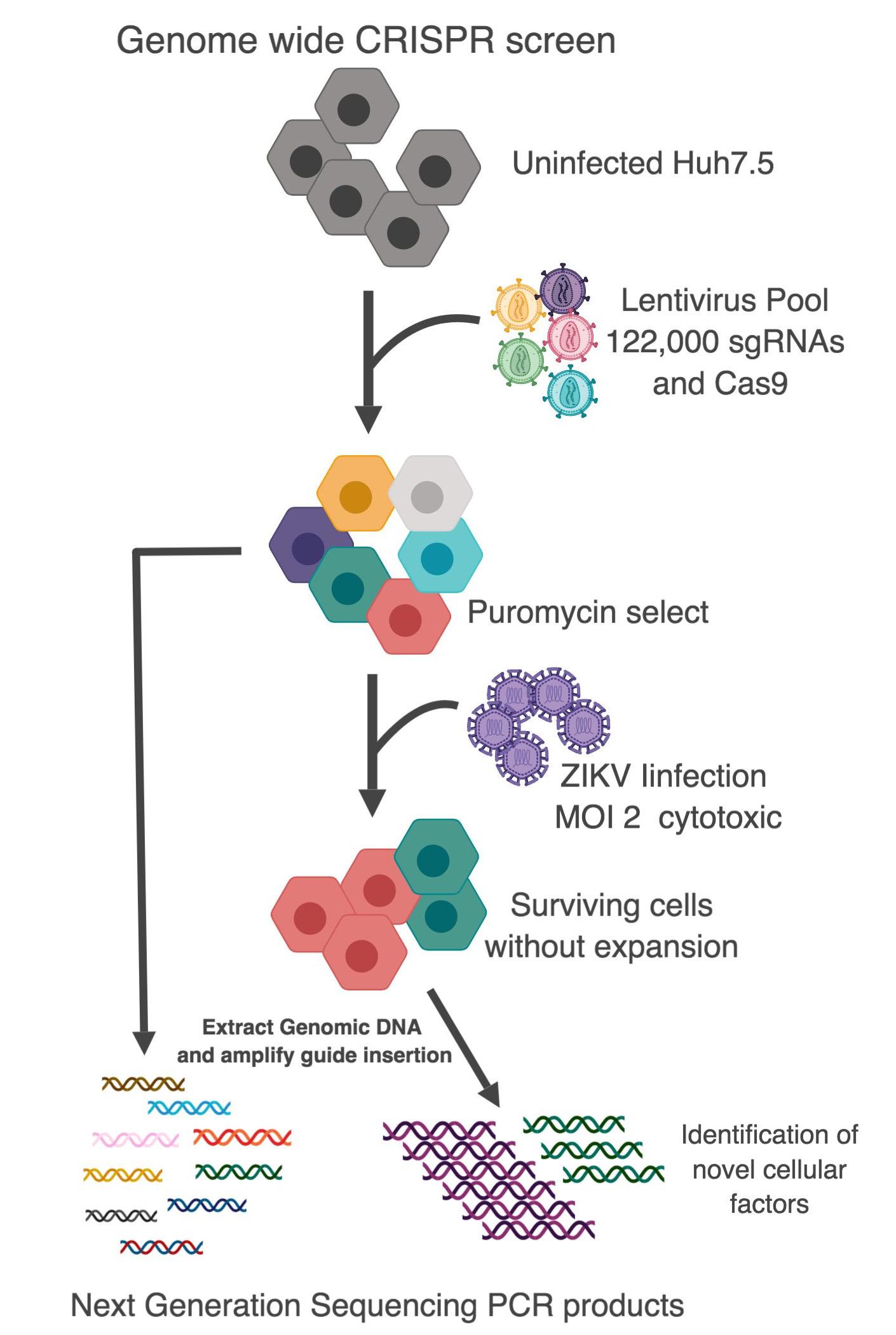
CRISPR/Cas9 technology is a powerful molecular tool to target to a specific gene for deletion and has revolutionised our ability to genetically manipulate cells.
Our laboratory has used a CRISPR/Cas9 genome wide screen in which guide-RNAs (approximately 122,000 delivered by lentiviral transduction) representing the complete human genome are introduced into cultured cells to selectively knock out cellular genes.
Using this technology followed by viral infection (ZIKV) we have identified novel essential host factors for viral replication such as the cellular protein RACK1. Interestingly, RACK1 is important for the replication of all Flaviviridae but not Ebola, MERS or DNA viruses suggesting selectivity in its action.
We are currently applying CRISPR/Cas9 genome wide screens to other viruses such as HCV and to identify novel genes that control virus replication in response to interferon stimulation.
Study with the Viral Pathogenesis Research Laboratory
Infection of the cell with a virus results in an early innate immune response in which the cell attempts to either remove the virus or control its replication.
Our laboratory is interested in the cellular response to viral infection and genes and signaling pathways that are turned on and how viruses can overcome this response. In particular we study the Flaviviridae family of positive strand RNA viruses such as hepatitis C (HCV) dengue and Zika virus.
Using a combination of cell culture, mouse and human primary tissue models coupled with the latest genomic and biochemical approaches we aim to define the cellular response to viral infection of these medical important viruses.

Supervisors
Associate Professor Michael Beard
Research area: Viral pathogenesis, Research Centre for Infectious Diseases
Recommended honours enrolment: Honours in Molecular and Biomedical Science